PlexPrime®
Novel nucleic acid amplification approach, creating amplicons distinctly different from the parent sequence. The combination of PlexPrime® and PlexZyme® technology enables multiplex mutation detection with high sensitivity and specificity.
Multiplex compatible
Detect multiple mutations in a single reactionVersatile
Flexibility for challenging sequencesHighly Specific
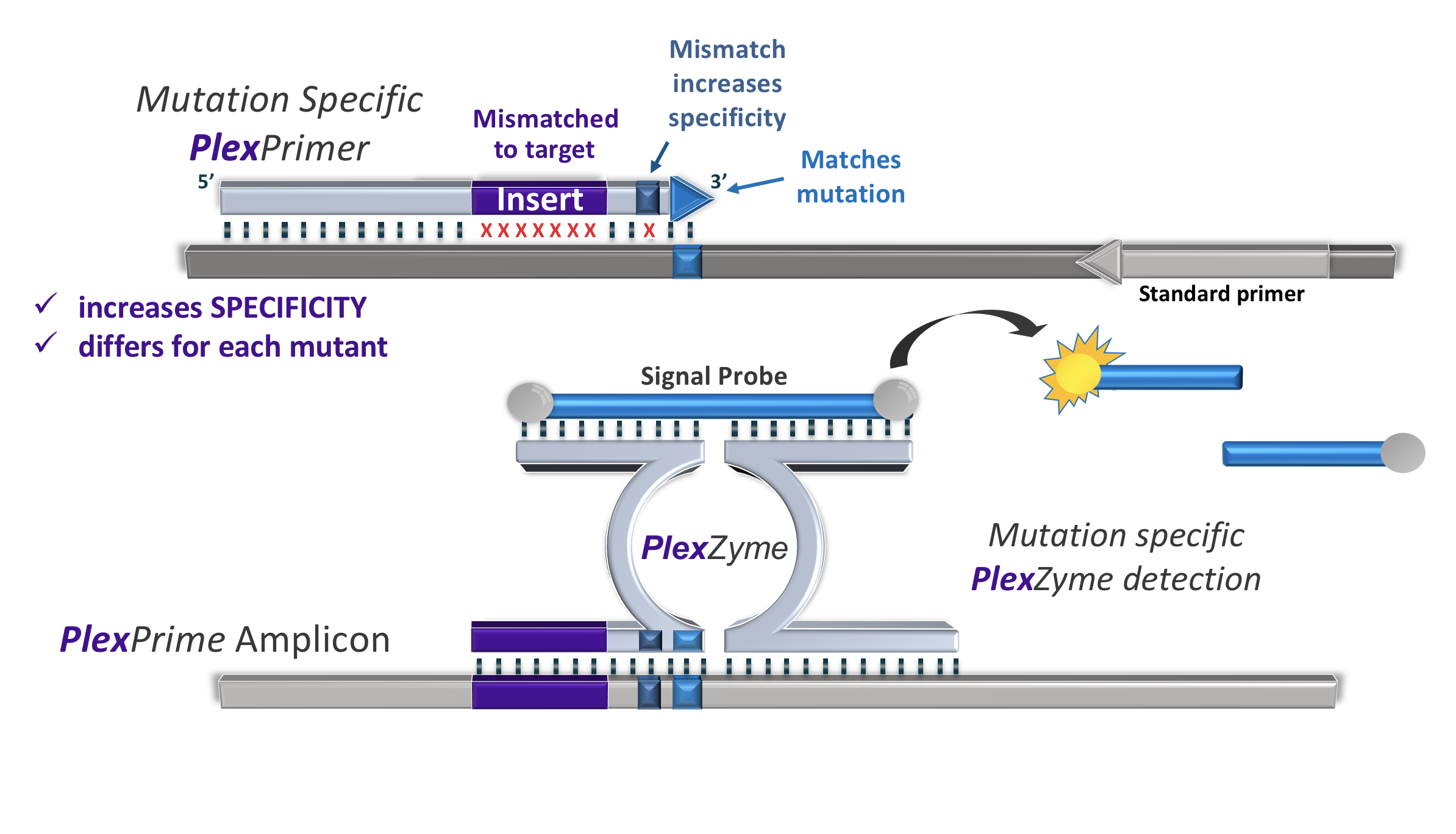
Increased stringency due to insert sequenceThe PlexPrime® primer is composed of three regions:
- 5’ region anchors the primer to a particular location in the target
- 3’ region selectively amplifies the target sequence of interest
- the Insert, acting as a bridging structure which both (a) inserts target-independent sequence into the resulting amplicon and (b) increases the selective pressure of the 3’ region
The unique feature of the PlexPrime® primer make it extremely suitable for detection and discrimination of mutations or Single Nucleotide Polymorphisms (SNPs) in a multiplex reaction or a pool of nucleic acid sequences containing non-conserved regions.
References
Tan L. Y., Walker S. M., Lonergan T., Lima N. E., Todd A. V. Mokany E. 2017. Superior Multiplexing Capacity of PlexPrimers Enables Sensitive and Specific Detection of SNPs and Cluster Mutations in qPCR. PLOS One January 23rd.
Link